643
 |
| Image credit: Rice University |
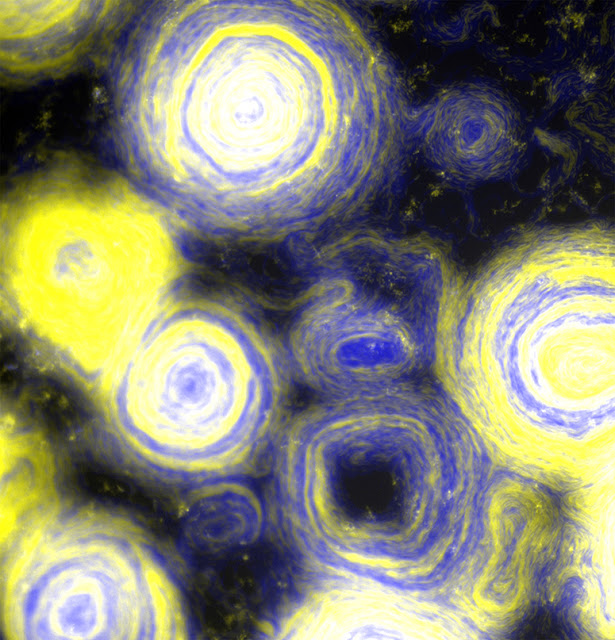
Scientists have figured out how to turn millions of predatory bacteria into spirals that look like Vincent Van Gogh’s The Starry Night. Myxococcus Xanthus is a species of myxobacteria that is capable of exhibiting different forms of self-organizing behaviour according to its environment and has been investigated as a model for social cooperation and bacterial gene regulation over the years. Bacteria are usually known to be selfish but M. Xanthus is different as it needs to find and recognize kin to survive.
Researchers uncovered a previously unknown activity while analyzing M. Xanthus mutants that overexpress two proteins that cells employ to distinguish near relatives: self-organization into millimetre-sized circles.
Oleg Igoshin, a professor of bioengineering at Rice University and senior scientist at Rice’s Center for Theoretical Biological Physics stated “When you overexpress that protein, you can see these circular aggregates emerge after four hours, and by 12 hours they take up the whole (petri dish).”
Igoshin’s research group has been collaborating with a microbiology group from the University of Wyoming, led by coauthor Daniel Wall and they have been conducting dozens of experiments to discover the genetic mechanism of the circular swarms.
M. Xanthus preys on other bacteria but lacks the internal organs that are required for digesting them further and hence they form familial packs to engulf and eat the victims. M. Xanthus uses a surface receptor called TraA and a companion protein called TraB to recognise kin, according to Wall and coauthor Pengbo Cao, who was then a graduate student in his lab and is now a postdoctoral research associate at the Georgia Institute of Technology.
Wall’s team established various mutant strains while researching TraAB’s mechanism, including ones that overexpressed TraAB and produced more of the protein than normal. They showed a tendency to create cell clusters after a few hours, Cao noted. While Wall’s team conducted microbiological tests, Igoshin’s team was instructed to develop a theoretical model that could explain the riddle.
“What’s interesting about our theory is that the only way we see these (circular aggregates) in our simulations is when we make the cells non-reversing. In normal wild-type cells, they go back and forth, back and forth, like a commuter train. The head becomes the tail and the tail becomes the head. And they do it every eight minutes or so,” said Igoshin.
The authors were able to confirm their hunch by running this situation using computational simulations. The conventional head-to-tail swarms became whirling swirls of cells as large as a millimetre or more with simple alterations to the TraAB proteins that resulted in their overexpression.
The result is a fascinating image of the microbial world, as well as a deeper knowledge of how millions of cells coordinate their movements.
The study was published in mSystems.
Contributed by: Mithun Sukesan
To ‘science-up’ your social media feed, follow us on Facebook, Twitter or Instagram!

